About this service
Using
HRaDis (
Homo
Repeats
and human
Diseases) service you can:
- Study relation of homo-repeats (single-amino-acid tandem repeats) to human diseases.
- Search for proteins with the given homo-repeat in the human proteome including the list of diseases and the GO annotations for these proteins.
- Study the coupling of different homo-repeats in one protein.
Eighteen known neurological diseases are associated with genetic abnormalities that elongate simple single letter motifs [1]. Thus, the presence of long (exceeding an acceptable value) polyglutamine and polyalanine repeats in proteins is associated with such diseases. Indeed, previous reports indicate that developmental diseases are associated with homo-repeat expansions such as poly-A (alanine): synpolydactyly type II (HOXD13), blepharophimosis (FOXL2), oculopharyngeal muscular dystrophy (PABPN1), infantile spasm syndrome (ARX), and holoprosencephaly (ZIC2) [2]. Expansion of polyalanine tracts causes at least 9 inherited human diseases, and the pathogenic mechanism of expanded polyalanine tracts contributing to the associated disease states remains poorly understood.
Expansion of poly-Q is implicated in several neurodegenerative diseases, including Huntington’s disease and several spinocerebellar ataxia types. It should be noted that the length of the polyQ repeat is critical to pathogenesis [3]. The cause of Huntington’s disease is the multiple insertion of a CAG codon that codes glutamine, in the IT_15 gene. The wild type genes of different people contain different numbers of the CAG repeats; however, if their number exceeds 36, the disease develops. Although, a 40 glutamine repeat is the normal allele present in the forkhead box P2 transcription factor, a protein that has not been found to be associated with a poly-Q disease [3].
Occurrence of homo-repeats in the protein sequence results in the increasing aggregation ability of the protein. The above mentioned data emphasize the importance of investigation of the functional role of amino acid homo-repeats.
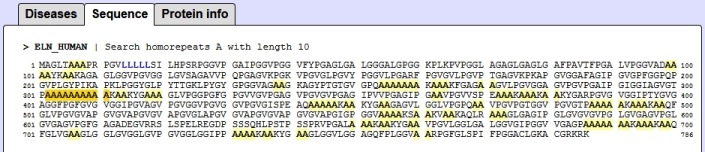
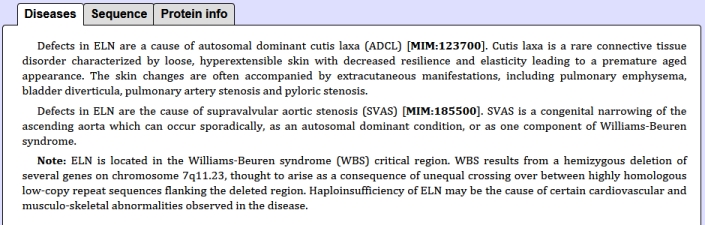
Example of polyalanin tracks in ELN_human protein and the list of diseases assoaciated with this protein:
- Jorda J., Xue B., Uversky V.N., Kajava A.V. 2010. Protein tandem repeats: The more perfect, the less structured. FEBS J. 277, 2673–2682.
- Mularoni L, Ledda A, Toll-Riera M, Albà MM. 2010. Natural selection drives the accumulation of amino acid tandem repeats in human proteins. Genome Res. 20:745–754.
- Robertson AL, Bate MA, Androulakis SG, Bottomley SP, Buckle AM. PolyQ: a database describing the sequence and domain context of polyglutamine repeats in proteins. Nucleic Acids Res. 2011 Jan;39(Database issue):D272-6.